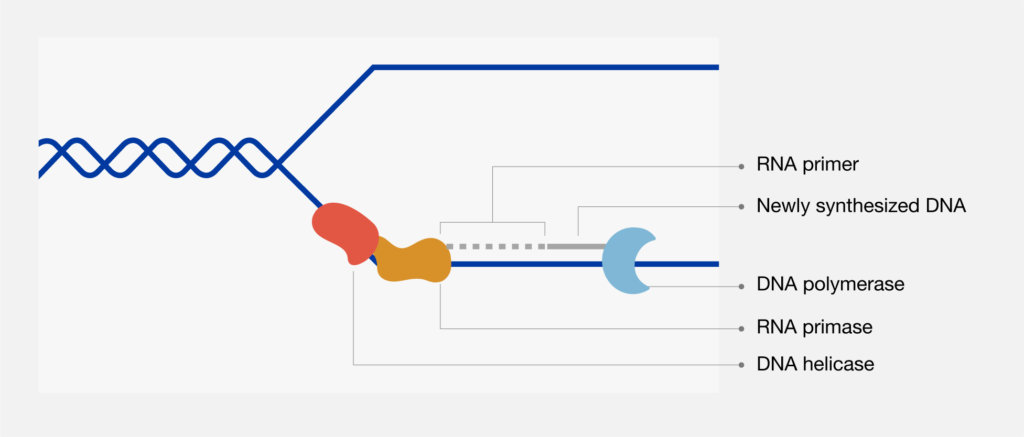
The principles of DNA replication were used by Sanger et al. in the development of the process now known as Sanger dideoxy sequencing. This process takes advantage of the ability of DNA polymerase to incorporate 2´,3´-dideoxynucleotides—nucleotide base analogs that lack the 3´-hydroxyl group essential in phosphodiester bond formation. Sanger dideoxy sequencing requires a DNA template, a sequencing primer, DNA polymerase, deoxynucleotides (dNTPs), dideoxynucleotides (ddNTPs), and reaction buffer.
Four separate reactions are set up, each containing radioactively labeled nucleotides and either ddA, ddC, ddG, or ddT. The annealing, labeling, and termination steps are performed on separate heat blocks. DNA synthesis is performed at 37°C, the temperature at which DNA polymerase has the optimal enzyme activity. DNA polymerase adds a deoxynucleotide or the corresponding 2´,3´-dideoxynucleotide at each step of chain extension. Whether a deoxynucleotide or a dideoxynucleotide is added depends on the relative concentration of both molecules. When a deoxynucleotide (A, C, G, or T) is added to the 3´ end, chain extension can continue. However, when a dideoxynucleotide (ddA, ddC, ddG, or ddT) is added to the 3´ end, chain extension terminates. Sanger dideoxy sequencing results in the formation of extension products of various lengths terminated with dideoxynucleotides at the 3´ end. The extension products are then separated by electrophoresis.
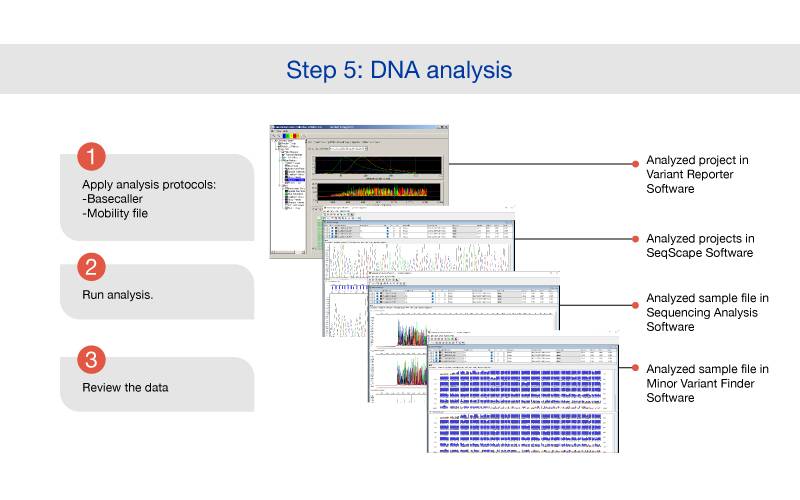
Applied Biosystems fluorescence-based cycle sequencing system is an extension and refinement of Sanger dideoxy sequencing. Applied Biosystems automated DNA sequencing generally follows this flow: template preparation, cycle sequencing, purification, capillary electrophoresis, and data analysis.

.jpg)